microglycodb_glycan_00083
| Glycan Structure | 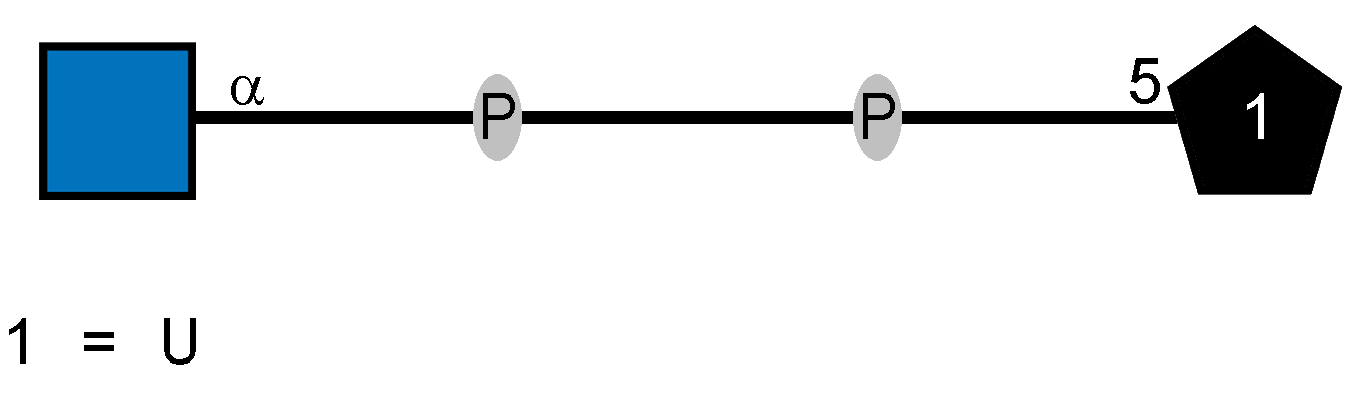 |
|---|---|
| Glycan Name | UDP-N-acetyl-alpha-D-glucosamine |
| CSDB Linear | Ac(1-2)aDGlcpN(1-P-P-5)xXnucU |
| IUPAC Condensed | |
| LINUCS | |
| WURCS | |
| Organism | Campylobacter jejuni |
Glycosyltransferases
External Links
| Gene ID | Gene No. | Gene Name | UniProt ID | Note | Reaction ID |
|---|---|---|---|---|---|
| microglycodb_gene_00081 | Cj1142 | neuC1 | Q0P9B2 | UDP-N-acetylglucosamine 2-epimerase / N-acetyl-mannosamine synthase | R00414 |
| microglycodb_gene_00090 | Cj1293 | pseB | Q0P8W4 | UDP-GlcNAc-specific C4,6 dehydratase / C5 epimerase | 26111 |
| microglycodb_gene_00139 | Cj1120c | pglF | Q0P9D4 | UDP-N-acetyl-alpha-D-glucosamine C6 dehydratase | 30823 |
| microglycodb_gene_00186 | Cj1039 | murG | Q9PNQ2 | UDP-diphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase, leads to the formation of Lipid II | 31227 |
| microglycodb_gene_00186 | Cj1039 | murG | Q9PNQ2 | UDP-diphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase, leads to the formation of Lipid II | 23195 |
| microglycodb_gene_00191 | Cj0858c | murA | Q9PP65 | Enolpyruvyl to UDP-N-acetylglucosamine as a component of cell wall formation | 18681 |